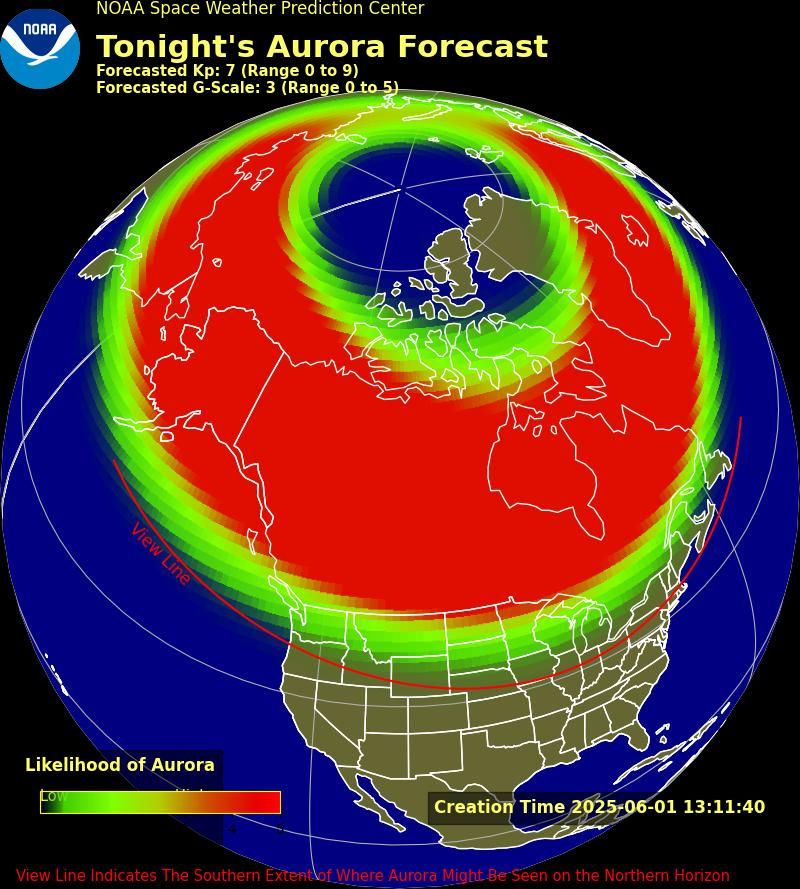
Wherever there are humans, there are microbes, too. Bacteria and fungi live all around us, in our homes, offices, industrial areas, the outdoors – even in space. People literally could not live without these tiny organisms, many of which are beneficial.
The trick is limiting potentially harmful ones, particularly in a contained environment such as a spacecraft. So from the launch of the very first module of the International Space Station, NASA has monitored its microbial community.
Because the station is an enclosed system, the only way that microbes get there is hitching a ride on the contents of resupply spacecraft from Earth and on arriving astronauts. The NASA Johnson Space Center Microbiology Laboratory puts a lot of effort into knowing which microbes ride along.
“We can’t sterilize everything we send into space, and don’t want to, but we do a lot to limit potential pathogens from making their way to the station,” says NASA microbiologist Sarah Wallace, Ph.D. “At launch, the cargo, food, vehicles, and crew members each have their own microbiome, or suite of microbes. When everything gets to the station, these microbiomes become part of the space station microbiome.”
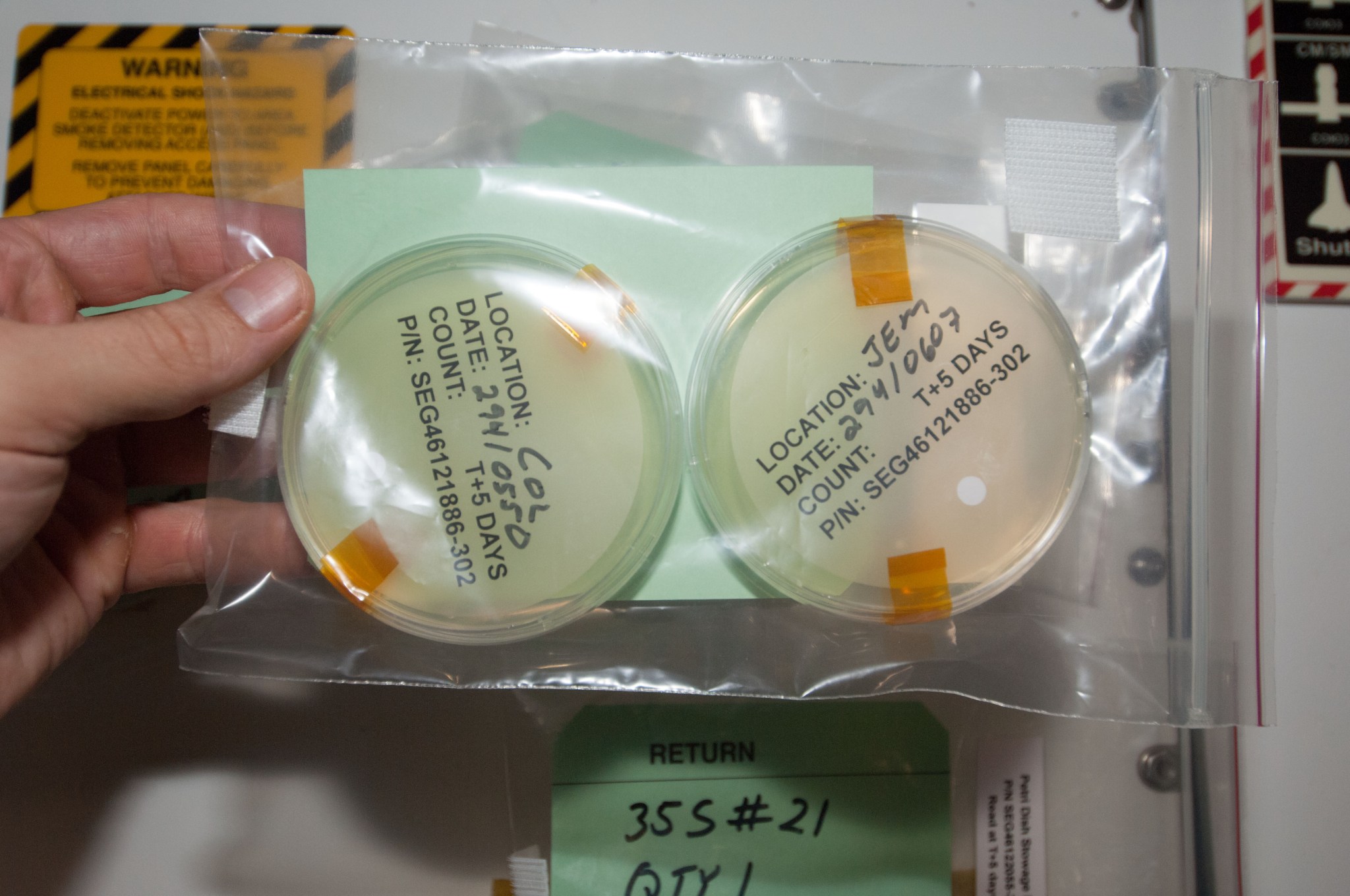
The lab uses the traditional method of culturing a sample in a growth medium, similar to Petri dishes from high school science class, to sample a portion of everything during packing for launch and the launch vehicles themselves. This sampling confirms that contamination control plans are working properly – essentially making sure the numbers of microbes remain low and that those present are the ones normally expected.

Then the lab continues monitoring after the vehicle, cargo, and crew arrive at the station. Crew members sample and culture microbes from the air, surfaces, and water on the station.
“It’s kind of a spot check to see how well housekeeping procedures are being implemented and how well the water system and the air filters are working,” Wallace says.
She calls the station’s water processing system “a phenomenal piece of engineering” that produces water much cleaner than most of us drink on Earth. In addition, the station itself is remarkably clean thanks to HEPA filters for the air and housekeeping practices for surfaces. “What microbes we see are really what we’d see if we looked at your home. In fact, we’ve done several studies comparing the station to a typical home and it is similar but usually cleaner,” she adds.
This monitoring over the lifetime of the orbiting lab has created a unique, long-term database that helps microbiologists know what to expect.
“Our requirements are two-fold, how much is there and what is there,” Wallace says. For years, the scientists didn’t know the ‘what’ until samples came back to ground. Now the equipment exists to perform direct swab-to-sequencer identification, eliminating the need to culture samples and return them to Earth. That equipment includes the miniPCR, a device that amplifies or makes many copies of a DNA strand using a process called polymerase chain reaction (PCR), and the MinION, a portable DNA sequencer. The Genes in Space 3 collaboration between Boeing and NASA paired these two platforms together, which led to the first identification of unknown bacteria off Earth.
NASA’s lab then conducted tests and confirmed that microbe identifications from the inflight process matched those determined on the ground down to the species level1.
“For the first time ever, we identified unknown microbes collected and cultured off Earth,” says Wallace. “We followed that up with the swab-to-sequencer, which lets us move away from culturing completely. We can swab a surface and sequence whatever is there.”
Subsequent work advanced the use of sequencing in space and later tests found that the culture-independent method showed the same microbial distributions as the standard culture-dependent method2. The swab-and-sequence method has been streamlined so that crew members can easily complete it in an extreme environment.
That is a critical capability for future missions to the Moon and Mars, both to continue to protect crew health and safety and to make sure that we do not contaminate other worlds. If explorers detect microbial life on another planet, they need to know whether it was already there or came from Earth.
Researchers also use the space station to conduct long-term microbial studies. The Microbial Tracking series studied what kinds of microbes are on the space station, both in the environment and in the astronauts’ bodies.
In addition to surveying the types of microbes present on the station, the lab studies whether those microbes could be harmful, as microgravity and radiation in space can render innocuous microorganisms potentially harmful and microbial behavior can change as the organisms adapt to the spaceflight environment.
So far, microbial issues on Earth far exceed any seen in space, Wallace says. “In addition to all the preflight monitoring, crew members are quarantined prior to launch. These steps were started back during Apollo missions and still are effective toward keeping our crews healthy.”
Because where people go, scientists want to know what microbes follow.
Citations
1 Burton AS, Stahl-Rommel SE, John KK, Jain M, Juul S, Turner DJ, Harrington ED, Stoddart D, Paten B, Akeson M, Castro-Wallace SL. Off Earth Identification of Bacterial Populations Using 16S rDNA Nanopore Sequencing. Genes. 2020 January 9; 76(11): 76 (https://www.mdpi.com/2073-4425/11/1/76)
2 Stahl-Rommel S, Jain M, Nguyen HN, Arnold RR, Aunon-Chancellor SM, Sharp GM, Castro CL, John KK, Juul S, Turner DJ, et al. Real-Time Culture-Independent Microbial Profiling Onboard the International Space Station Using Nanopore Sequencing. Genes. 2021; 12(1):106. (https://www.mdpi.com/2073-4425/12/1/106)